Abstract
Nitrogen efficiency (Neff; milk N/N intake) in dairy cows is limited and most of the consumed N is excreted in manure. Despite the crucial role of the gastrointestinal microbiome on N metabolism, associations between bacterial communities at different sections and Neff are not fully elucidated. Enhanced understanding of host-microbiome interactions can provide insights to improve Neff in dairy cows. Twenty-three Holstein cows were selected, and their Neff were determined using a N balance approach. From the cohort of cows, six cows were classified as low Neff and five cows as high Neff and their rumen and fecal bacterial communities were profiled using amplicon sequence variants (ASV) based on 16S rRNA gene sequencing. Then, relationships between differentially abundant bacterial features and Neff were evaluated. Neff in low and high cows averaged 22.8 and 30.3%, respectively. With similar N intake, high Neff cows wasted less N in manure compared to low Neff cows (P < 0.01, 11.0 ± 0.59 vs 14.3 ± 0.54 g of N/kg of milk). Rumen fermentation and plasma profiles were similar between Neff groups, but for plasma Gln which was greater (P = 0.02) in high compared to low Neff cows. In both rumen and feces, the phylogenetic composition of the bacterial communities was similar (P ≥ 0.65) between Neff groups, but differences were observed at the species -level (amplicon sequence variants). In the rumen, differentially abundant species from the genus Prevotella showed strong positive correlations with Neff, whereas in feces, differentially abundant species from the class Clostridia showed strong negative correlations with Neff. Our results revealed that Holstein cows with divergent Neff display distinctive bacterial community structure at the species-level in both the rumen and feces. Strong correlations between differentially abundant species and Neff in both sample sites, support the importance of the rumen bacterial composition on productive responses and suggest a more relevant role of the hindgut microbiome. Targeting both pre- and post-gastric bacterial communities may provide novel opportunities to enhance Neff in dairy cows.
Introduction
Dairy cow enterprises are important contributors to global food security supplying ~ 80% of the total milk1. Over the past decades, intensification of the dairy sector has resulted in fewer dairy farms with a greater herd size and milk output2. Scale-up of dairy operations decreases the cost of milk production, but also results in challenges with nutrient management3. Generally, dairy cows exhibit a low N efficiency (Neff; milk N/N intake) and ~ three-fourths of the consumed N is excreted in manure which is of environmental concern4,5. Feed is the greatest cost of milk production and protein supplements are main components in dairy diets, thus improvement in Neff will assist farm finance and reduce N waste6,7. Reported Neff of 35% under commercial settings with no adverse impacts on milk yield and composition indicates an opportunity for improvement8,9.
The gastrointestinal microbiota serves as an intermediary between dietary inputs and animal end products. In ruminants, the relevance of the rumen microbiota in N metabolism is well recognized as a main portion of the consumed protein is degraded by rumen microbes and microbial crude protein (CP) represents more than 50% of the metabolized protein10,11. However, only until recently has the structure of the rumen microbial community gained greater notice. Studies have identified subsets of rumen microbes that can predict productive traits in both dairy and beef cattle12,13. These results suggest that specific microbial members can shift rumen fermentation to yield compounds that are of more benefit to the animal. For instance, repeated inoculation with bison rumen contents improved the total tract N digestibility of heifers fed a barley straw diet and this response was correlated with shifts in the bacterial families Christensenellaceae and BS11 gut group and total protozoa14. The microbial community of the hindgut also has the capacity to degrade N sources and plays a role in N recycling15. Similar to the rumen, associations between the hindgut microbiome and feed efficiency and milk traits have been reported16,17. Evaluations of the relationships between the microbial communities of the rumen and hindgut and Neff are needed to provide insights about host-microbiome interactions to improve the conversion of N into high quality milk protein.
The objectives of this study were to characterize the rumen and fecal bacterial communities of dairy cows with divergent Neff, identify bacterial features that differed between Neff groups, and evaluate the extent of the relationships between the identified differential bacterial features and Neff. We hypothesized that for both the rumen and hindgut, the global profile of the bacterial community will be similar between divergent Neff groups, but differences will be observed for specific groups, and these will show strong correlations with Neff.
Results
N efficiency phenotype
Based on the Neff phenotype selection criteria, 6 cows were classified as low Neff and 5 cows were classified as high Neff (Supplementary Table S1). In these cows, Neff ranged from 20.7 to 34.1% and significantly differed (P < 0.01; Table 1) between the low and high phenotypes averaging 22.8 and 30.3%, respectively.
N balance in low and high Neff cows
Production and N partitioning responses between low and high Neff cows are shown in Table 1. The DMI were similar (P = 0.27) between the low and high Neff cows and averaged 26.7 kg/day, but high Neff cows produced 6.6 kg/day more (P < 0.01) milk than low Neff cows. Between Neff groups, milk composition did not differ (P ≥ 0.26) and averaged 3.00 and 3.84% for protein and fat; respectively, and SCC was also similar (P = 0.06) and averaged 58.1 cells/mL. In line with DMI and milk yield responses, N intake was similar (P = 0.27) between Neff groups, but milk N, determined from true protein, was greater (P = 0.03) in high Neff compared to low Neff cows (199 vs 162 g/day, respectively). Partitioning of N in urine and feces was similar (P ≥ 0.20) between Neff groups and consequently manure N (urinary N + fecal N) did not differ (P = 0.29). However, when manure N was expressed per kg of milk produced, low Neff cows wasted more N in urine (P = 0.04) and feces (P < 0.01) than high Neff cows, thus manure N was also greater (P < 0.01; 14.3 vs 11.0 g of N/kg of milk, respectively).
Ruminal fermentation
Ruminal fermentation variables are summarized in Table 2. The rumen environments were comparable between the low and high Neff cows. No differences were observed in pH (P = 0.63), NH3 concentration (P = 0.37), and total VFA concentration (P = 0.88). In addition, the molar proportions of acetic (P = 0.34), propionic (P = 0.35), butyric (P = 0.99), and valeric acid (P = 0.33) were similar between Neff groups.
Plasma AA profile
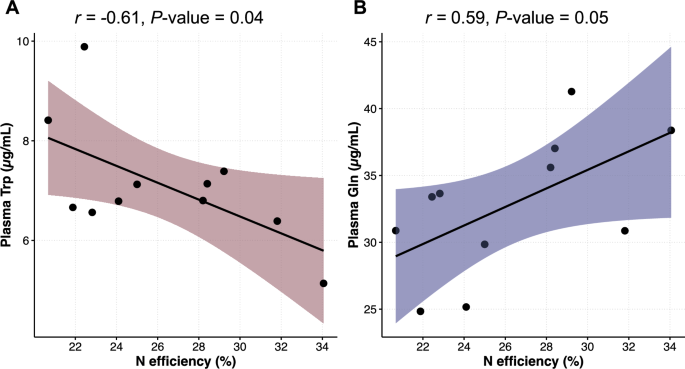
Plasma concentration of essential AA (EAA; Arg, His, Ile, Leu, Lys, Met, Phe, Thr, Trp, Val) did not differ (P ≥ 0.06) between Neff groups (Supplementary Table S2). Likewise, plasma concentration of total EAA (TEAA) did not differ (P = 0.14) and averaged 142 µg/mL. For non-essential AA (NEAA), plasma concentrations between Neff groups were similar (P ≥ 0.15) but for Gln (P = 0.02) which was greater in high Neff compared to low Neff cows (36.6 vs 29.6 mg/mL). To further evaluate relationships between plasma concentration of individual AA and Neff, Pearson correlations were determined. A moderate negative correlation (r = − 0.61, P = 0.04) was determined for Trp, while a moderate positive correlation (r = 0.59, P = 0.05) was determined for Gln (Fig. 1). For the remaining AA, correlations with Neff were not significant (P ≥ 0.10; Supplementary Table S3).
Sequencing information
Across quality-filtered, rarefied samples, 2250 bacterial ASV were identified. Rarefaction curves from both sample site and Neff phenotype tended to plateau indicating an adequate sequencing depth for bacterial community analyses (Supplementary Fig. S1). In addition, the Good’s coverage index estimated that at least 97% of the bacterial community was characterized across samples. Thus, analyses in this study accurately evaluated dominant members of the bacterial populations in the rumen and feces from low and high Neff cows.
Sample site
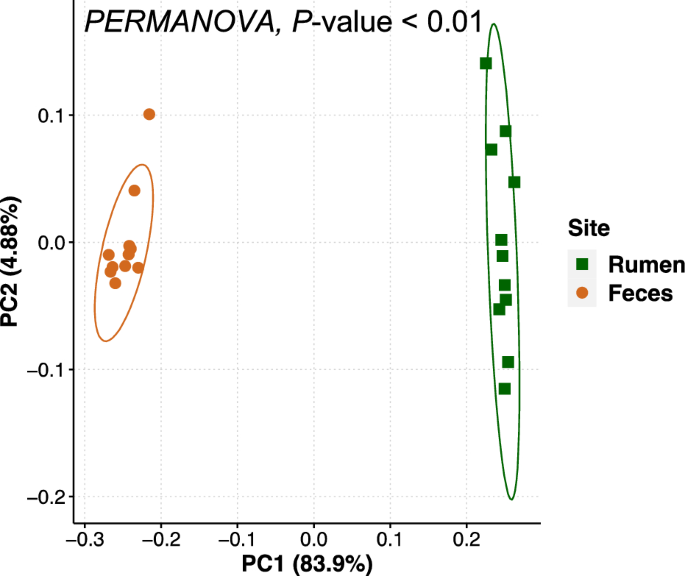
The bacterial communities differed by sample site (P ≤ 0.01) which was clearly visualized in the PCoA plot (Fig. 2). In the rumen, Bacteroidetes, Proteobacteria, and Firmicutes were the most abundant phyla and accounted for 74.3% of the total quality-filtered reads. Other predominant phyla were Spirochaetota (8.4%), Verrucomicrobiota (6.2%), Fibrobacterota (6.0%), Cyanobacteria (2.7%) and Patescibacteria (1.3%). In the fecal samples, the most abundant bacterial phyla were Bacteroidetes and Firmicutes, which accounted for 88.3% of the total quality-filtered reads. Other predominant phyla in the fecal samples were Spirochaetota (5.2%), Proteobacteria (2.9%), and Actinobacteriota (1.5%). The taxonomic classifications of the rumen and fecal bacterial communities across ranks are shown in Supplementary Fig. S2.
Diversity analysis
α-Diversity
The alpha metrics used to describe the bacterial community richness (observed ASV’s), evenness (Pielou’s index) and diversity (Shannon index) from the rumen and feces in high and low Neff cows are presented in Table 3. In the rumen bacteriome, high Neff cows presented a greater richness than low Neff cows (P = 0.04). However, alpha metrics for evenness (P = 0.86), and diversity (P = 0.47) were similar between both groups. In the fecal bacteriome, alpha metrics for richness (P = 0.72), evenness (P = 0.99), and diversity (P = 0.72) were similar.
β-Diversity
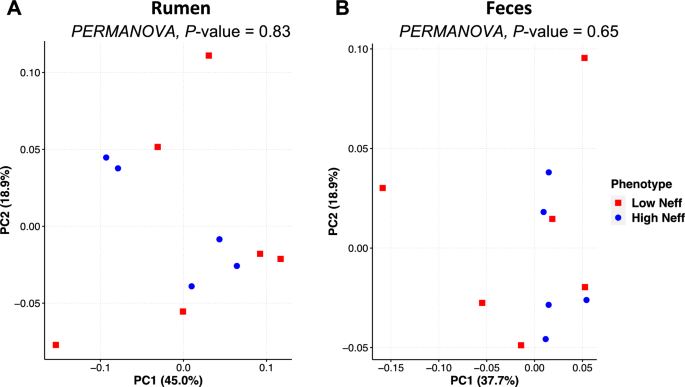
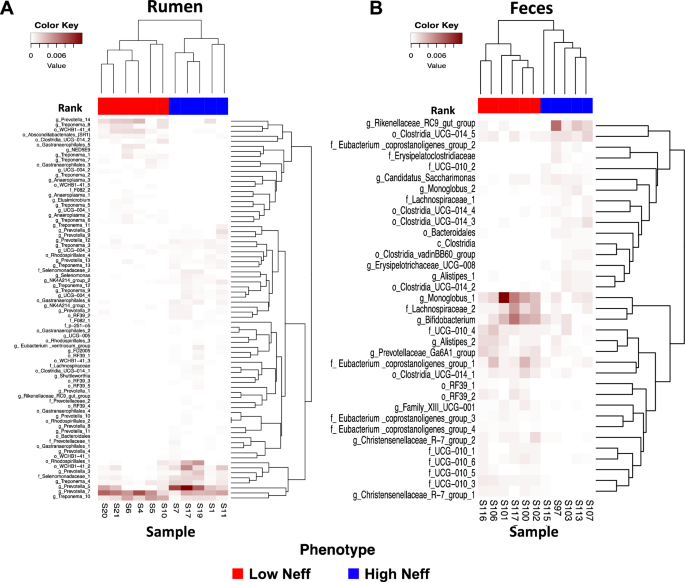
The phylogenetic composition of the bacterial communities in both the rumen and feces were similar between Neff groups (P ≥ 0.65) and this was reflected by no clear grouping of samples in the PCoA plot (Fig. 3). At the ASV-level, 76 ruminal and 35 fecal differentially abundant ASV were identified using LEfSe between low and high Neff cows (Fig. 4). Hierarchical clustering showed distinct groups of differentially abundant ASV between Neff phenotypes in both the rumen and feces. In the rumen, differentially abundant ASV mainly belonged to the genera Prevotella (14 ASV) and Treponema (13 ASV); whereas in the feces, differentially abundant ASV mainly belonged to the order Clostridia UCG-014 (5 ASV) and to the families UCG-010 (6 ASV) and Eubacterium coprostanoligenes group (4 ASV).
Correlations between differentially abundant bacterial ASV and Neff
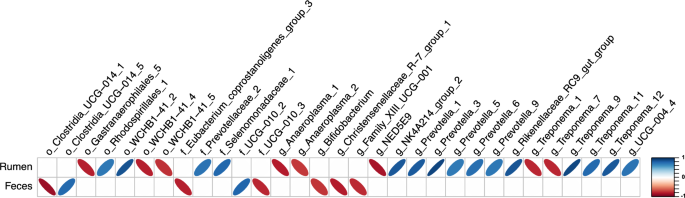
Correlations were performed to evaluate the relationships between the relative abundance of differentially abundant bacterial ASV from the rumen and feces and Neff (Fig. 5). In the rumen, ASV from the order Rhodospirillales, the families Prevotellaceae and Selenomonadaceae, and the genera NK4A214 group, Prevotella, Rikenellaceae RC9 gut group, and UCG-004 had strong positive correlations with Neff, whereas ASV from the order Gastranaerophilales and the genera Anaeroplasma and NED5E9 had strong negative correlations with Neff. ASV belonging to the order WCHB1 and the genus Treponema had both positive and negative strong correlations with Neff. Moderate correlations were also identified between 24 ruminal ASV and Neff where ASV from the genus Prevotella had positive correlations and ASV from the genus Treponema mainly had negative correlations (Supplementary Table S4). In feces, 8 differentially abundant ASV were identified to have strong correlations with Neff and the majority (6 ASV) had negative associations and belonged to the order Clostridia UCG-014, the families Eubacterium coprostanoligenes group and UCG-010, and the genera Bifidobacterium, Christensenellaceae R-7 group, and Family XIII UCG-001. In addition, 9 ASV had moderate positive and 6 ASV had moderate negative correlations with Neff (Supplementary Table S5).
Discussion
In lactating cows, N intake is a main factor affecting the partition of N between milk and manure5,18. As N intake increases, there is decrease in N efficiency18 and consequently, manure N increases19,20. In the current study, Neff groups had a similar N intake, but N partitioned towards milk protein was greater in high Neff compared to low Neff cows. This response was driven by milk yield since milk protein concentration was similar between groups. Similar observations were reported in a study under commercial conditions where herds averaging 30% Neff had higher milk yield and similar milk composition compared to herds averaging 22% Neff8. For manure N, high Neff cows excreted less urinary and fecal N per kg of milk than low Neff cows. Nitrogen excreted in manure represents an economic loss and an environmental burden, particularly urinary N which is rapidly lost through volatilization21,22.
Rumen microbial fermentation contributes to feedstuff degradation in ruminants. This process provides microbial CP and VFA which are the main sources of AA and energy to the animal, respectively11,23. Moreover, the rumen fermentation profile can impact production responses in dairy cows24. In ruminant nutrition, the portion of dietary CP that is degraded by rumen microbes is defined as rumen-degradable, whereas the portion that bypasses the rumen is defined as rumen-undegradable. Across feeds, rumen-degradable protein ranges from 30 to 85% and its degradation by rumen microbes yields peptides, AA, and NH3 that are used for microbial CP synthesis10,25. However, degraded CP not incorporated into microbial CP is converted to NH3, absorbed through the rumen wall and eventually disposed as urea in the urine. Given the importance of rumen microbial fermentation on N metabolism, the rumen fermentation profiles between Neff groups were evaluated. Similar rumen environments were observed between high and low Neff groups. In contrast, a study using goats found lower concentrations of NH3 and propionic acid in the high Neff group compared to the low Neff group and associated these responses with increased microbial CP synthesis26. Discrepancies between studies can potentially be attributed to differences in ruminant species and diet27,28. In general terms, plasma concentration of AA reflects the interplay between AA pool inputs and outputs. Meta-analytic research suggests a linear relationship between the predicted individual duodenal EAA flows and their respective plasma concentration in lactating dairy cows29. We observed plasma concentration of Gln to be greater in high Neff compared to low Neff cows and a moderate positive correlation between plasma Gln and Neff. Factors driving the difference in plasma Gln were not clear since supply was most likely similar due to similar DMI between Neff groups and Gln generation from the urea cycle might not be a contributor due to similar plasma urea between Neff groups30. We also observed a moderate negative correlation between plasma Trp and Neff and this potentially might reflect imbalances of AA as Trp supply increases.
Studies have shown that the microbial community structure in the rumen and lower gut is relevant to animal productivity12,17. Commonly, no differences are observed between extreme phenotypes at the community level but at particular taxonomic levels31,32. In the present study, beta diversity was similar between Neff groups, but differentially abundant ASV were identified. In the rumen, Prevotella is an abundant and core genus33,34 and in this study the relative abundances of the majority of the differentially abundant ASV belonging to Prevotella were greater in high Neff compared to low Neff cows. In addition, these Prevotella species presented strong positive correlations with Neff. Regarding rumen N metabolism, Prevotella species were correlated to branched chain AA biosynthesis in high milk protein yield cows suggesting greater microbial CP synthesis35. Also, Prevotella ruminicola and microbial CP concentration had been reported to be greater in high Neff compared to low Neff goats26. In the current study, ASV belonging to the Treponema genus were also identified to differ between Neff groups. The Treponema genus is another member of the core group of the rumen microbiome36 and species of this genus contribute to the ruminal fatty acid pool37. Mixed results were observed for microbes from the Treponema genus as around half of the differentially abundant ASV were greater in the low Neff group and the remainder were greater in the high Neff group. Likewise, 2 strong negative and 3 strong positive correlations were observed in ASV from the Treponema genus.
The profile of the main phyla in the hindgut is similar to that of the rumen, but the relative abundances and the microbes within these groups vary considerably38. Previous studies evaluating the bacterial profile from the hindgut in cattle have shown differences in the abundance of community members between contrasting feed efficiency animals and have also reported associations with productive traits such as milk protein17,39. The latter suggests that it is relevant to identify bacterial groups across the different sections of the gastrointestinal tract that impact nutrient efficiency. In this study, mainly strong negative correlations were identified between fecal bacterial ASV and Neff and in general these ASV belonged to the class Clostridia. Microbes within the Clostridia taxon are obligate anaerobes that commonly reside within the gastrointestinal tract of farm animals and can ferment plant polysaccharides40. However, multiple species can produce toxins causing enteric diseases41. Likewise, a negative strong correlation between the family Lachnospiraceae (within the class Clostridia) and Neff was reported in the colon of goats26.
Conclusions
In ruminants, the gastrointestinal microbiome has a central role in N metabolism; however, differences in the bacterial communities between divergent Neff groups are unclear. We evaluated the bacterial community profiles of the rumen and hindgut between low and high Neff dairy cows. In both rumen and fecal samples, we identified clusters of differentially abundant bacterial features that were distinct between low and high Neff cows. In the rumen, differentially abundant species from the genus Prevotella showed strong positive correlations with Neff; whereas in feces, differentially abundant species from the class Clostridia showed strong negative correlations with Neff. We also report a positive correlation between plasma Gln and Neff. In addition to the rumen microbiome, the microbiome harbored in other sections of the gastrointestinal tract can impact the efficiency N is converted into milk protein.
Methods
Animals, housing and management
All experimental procedures involving animals were performed under the approval of the Mississippi State University Institutional Animal Care and Use Committee (IACUC #19-581). The study complies with ARRIVE guidelines and all methods were performed in accordance with the relevant guidelines and regulations. A 28-day trial was conducted with 23 mid-lactation (164 ± 37.7 days in milk; mean ± SD) Holstein cows at 4.8 ± 0.8 years of age with 633.5 ± 49.8 kg of body weight (BW), 2.90 ± 0.40 of body condition score and producing 37.7 ± 6.5 kg/day of milk. The first 14-day served as an adaptation period to the Calan Broadbent Feeding System (American Calan Inc., Northwood, NH, USA) to record individual intake. The diet was formulated to meet or exceed the requirements of a lactating cow11 and comprised of 50% forage and 50% concentrate (dry matter (DM) basis; Supplementary Table S6). At the end of the adaptation period, cows were able to access their assigned feed bin without assistance and exhibited stable intake and milk production patterns (Supplementary Fig. S3). The subsequent 14-day served as the collection period of samples.
All cows were housed in a free-stall barn within the same pen with a stocking density of 96% and were under the same management protocol during the study. Cows were provided 2 ×/day (0600 and 1800 h) fresh total mixed ration (TMR) for ad libitum consumption and had free access to water troughs. The amount of TMR offered to each cow was adjusted daily to maintain approximately 10% refusals (as is basis) based on individual refusals from the previous day. Intake was calculated as the difference between the amount of feed offered and refused. The cows were milked 2 ×/day (0300 and 1500 h) in a double 8 parallel milking parlor at which the animals were moved as a group. No health complications were recorded, and all cows completed the study. Following the completion of the study, cows were returned to the standard farm management protocols.
Nitrogen balance
Feed and milk composition
TMR and refusals were sampled on days 21, 22, 27 and 28 and stored at − 20 °C. Composited samples of TMR and refusals were sent to Cumberland Valley Analytical Services (Waynesboro, PA) for DM, CP [42; method 990.03], neutral detergent fiber (NDF)43, and acid detergent fiber (ADF) [42; method 973.18] analyses. Individual milk yield was recorded daily throughout the experiment (Westfalia Surge Metatron 12 Milk Meter; GEA Farm Technologies, Oelde, Germany). From day 24 to 26, milk samples were collected from each milking using Waikato milk meters (Waikato Milking Systems, Horotiu, New Zealand) and stored at room temperature in plastic vials with a tablet preservative containing 8 mg bronopol and 0.30 mg natamycin (Broad Spectrum Microtabs II; Advanced Instruments, Norwood, MA) before shipping to Mid-South Dairy Records (Springfield, MO, USA) for true protein and fat content, and somatic cell count (SCC) analyses by Fourier-transform infrared spectroscopy (Bentley FTS/FCM; Bentley Instruments, Chaska, MN, USA). Calculations for N intake and milk N were as follows:
where N intake = g/day, intake and refusals = kg/day, and diet and refusals CP = proportion of CP in the sample.
where Milk N = g/day, milk yield = kg/day, and milk true protein = proportion of true protein in the sample.
Fecal and urinary N
To account for diurnal variation, fecal and urine samples from each cow were concomitantly collected on day 26 at 0000, 0900 and 1800 h, on day 27 at 0300, 1200 and 2100 h, and on day 28 at 0600 and 1500 h (n = 8). This sampling scheme allowed to collect samples representing 3-h intervals in a 24-h period. Fecal samples (~ 200 g) were collected via fecal grab and stored in inert polyethylene cups at − 20 °C. Spot urine samples (~ 450 mL) were collected via vulva stimulation and immediately after each collection, an aliquot of 40 mL was acidified to pH < 3 with 3 mL of 10% sulfuric acid (H2SO4) and stored at − 20 °C. An additional 40 mL aliquot of urine without acid was stored at − 80 °C. Fecal samples were thawed and composited by cow and then samples were air dried at 60 °C for 48 h and subsequently ground through a 2 mm screen using a Wiley mill (Arthur H. Thompson Co., Philadelphia, PA). Ground fecal samples were analyzed for DM, N [42; method 2001.11] using a Kjeltec Auto Sampler System 1035 Analyzer (Tecator Inc., Höganäs, Sweden), and NDF [42; method 2002.04] and ADF [42; method 973.18] using an Ankom 200 Fiber Analyzer (Ankom Technology, Macedon, NY, USA). Acidified urine samples were thawed, composited by cow, and shipped to Cumberland Valley Analytical Services (Waynesboro, PA) for N analysis [42; method 990.03]. Non-acidified urine samples were thawed, composited by cow, and analyzed for creatinine concentration using the creatinine colorimetric assay (Cayman Chemical Co., Ann Arbor, MI, USA) according to the manufacturer’s protocol. Urinary creatinine was used as a marker to estimate urine volume as44:
where Urine volume = L, creatinine excretion = 29 mg/kg of BW, BW = kg, and creatinine = mg/L. Urinary N losses were then calculated as the product of urine volume and urine N concentration (acid adjusted).
Apparent nutrient digestibility was estimated using the nylon bag techniques with indigestible acid detergent fiber (iADF) as the internal marker45. Samples of ground TMR and feces (from each cow) were weighed in triplicates into 5 × 10 cm in situ bags with 50 ± 10 μm pores (R510; Ankom Technology, Macedon, NY, USA) and incubated in the rumen of two cannulated cows for 12 days. The canulated cows were under the same experimental diet. After incubation, the in situ bags were immediately placed on ice and then gently rinsed with running tap water to remove attached particles and stop microbial activity. Digested samples were later dried at 60 °C for 48 h and residues were analyzed for iADF following the procedure specified for the fecal grab samples. Apparent nutrient digestibility (%) was calculated as follows:
Fecal N was then calculated as the difference between N intake and digestible N.
For each cow, Neff was calculated as8,46:
where N efficiency = %, milk N = g/day, and N intake = g/day. Cows with 0.5 standard deviation (SD) below and above the Neff mean were classified as low (Neff < mean + 0.5 × SD) and high (Neff > mean + 0.5 × SD) Neff, respectively26,47.
Bacterial community composition and rumen fermentation
Rumen and fecal sampling
On day 27, rumen samples were collected at 0500 h (before morning feeding) using a gastroesophageal apparatus that consisted of a reinforced vinyl tube coupled on one end to a metal strainer and on the other end to a 500 mL sterile collection flask that was connected to a vacuum pump (model 1HAB25BM100X; Gast, Benton Harbor, MI, USA). Rumen sampling and preparation of rumen samples for bacterial community profiling were done as described by Paz et al.48. Immediately after sampling, pH was measured using a portable pH meter (handheld pH meter, Oakton, Vernon Hills, IL) by direct insertion into an aliquot of 50 mL of rumen fluid. For ammonia (NH3) and volatile fatty acids (VFA) analyses, aliquots of rumen fluid (45 mL each) were filtered through 4 layers of cheesecloth and stored at − 20 °C. Following rumen sampling for each cow, approximately 200 g of feces were collected via fecal grab for bacterial community profiling. Fecal samples were placed in sterile plastic containers and stored at − 80 °C until further analysis.
DNA extraction, library preparation, and sequencing
Genomic DNA was extracted from rumen and fecal samples as described by De La Guardia-Hidrogo and Paz49. Amplified DNA integrity was checked through gel electrophoresis (1% agarose gel) and concentration and purity (260-to-280 nm ratio) were determined using the NanoDrop One Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Bacterial amplicon libraries of the V4 region from the 16S rRNA gene were prepared as described by Kozich et al.50. Briefly, each 20 μL PCR amplification reaction contained 0.5 μL Terra PCR Direct Polymerase Mix (0.625 Units; Takara Bio, Mountain View, CA, USA), 7.5 μL nuclease-free, sterile water, 10 μL 2 × Terra PCR Direct Buffer (Takara Bio, Mountain View, CA, USA), 1 μL indexed primers (10 μM) and 1 μL DNA (25–70 ng DNA). The cycling conditions included an initial denaturation of 98 °C for 3 min, followed by 25 cycles of 98 °C for 30 s, 55 °C for 30 s, and 68 °C for 45 s, and a final extension of 68 °C for 4 min. The resulting amplicons were normalized (1–2 ng/μL) using the SequalPrep™ Normalization Plate Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Normalized amplicons were pooled (10 μL/sample) and libraries were purified using the MinElute PCR Purification Kit (Qiagen Inc., Valencia, CA, USA) as described by the manufacturer. Finally, libraries were sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA) at the Institute for Genomics, Biocomputing, and Biotechnology (Mississippi State, MS, USA). Raw sequences are available at the NCBI Sequence Read Archive (SRA) under the accession no. SRP400507.
Ruminal fermentation
Prior to analysis, samples for NH3 and VFA were thawed. For NH3 concentration, a portable meter (Orion meter model 290; Thermo Scientific Inc., Waltham, MA, USA) coupled to an NH3 ion selective electrode (Orion 9512 ammonia sensing electrode; Thermo Scientific Inc., Waltham, MA, USA) was used. For VFA profile, 1 mL of filtered rumen fluid was mixed with 0.2 mL of an internal standard (2-mg/mL 2-ethyl butyric acid in 25% meta-phosphoric acid (w/v)). Subsequently, samples were centrifuged at 18,000×g for 20 min, and the resulting supernatant was analyzed using a 7890A gas chromatography system equipped with a DB-FATWAX Ultra Inert column (30 m × 0.25 mm, 0.25 µm), a 5975C inert XL MSD with triple-axis mass detector, and a 7693 series autosampler (Agilent Technologies, Santa Clara, CA, USA). Ionization was performed in an electron impact mode at 70 eV and a selected ion monitoring mode was used to acquire ion abundance. Volatile fatty acids were quantified by an internal standard calibration with authentic VFA standards.
Plasma amino acids (AA)
To evaluate plasma AA profiles between Neff groups, blood samples were collected on day 27 at 1800 h via venipuncture of the coccygeal artery. Samples were collected into evacuated tubes containing K2EDTA (BD Vacutainer; Becton Dickinson and Co., Franklin Lakes, NJ, USA). Immediately after collection, samples were placed on ice and plasma was separated as described by Paz and Kononoff51. Plasma samples were stored at − 20 °C and later shipped to the Agricultural Experimental Station Chemical Laboratories (Columbia, MO, USA) for free plasma AA analysis using a Hitachi L-8900 AA analyzer (Hitachi Ltd., Tokyo, Japan)52,53.
Bioinformatics analysis
Detailed information of the complete bioinformatic analysis containing the metadata file and sequential R markdown files with the scripts and commands used in this study can be found at https://github.com/pazlabgit/Neff_holstein_pipeline. Briefly, demultiplexed sequences were analyzed using QIIME 2 v2020.8.054. The denoising process included an initial quality filtering based on quality scores (q2-quality-filter plugin)55 followed by the Deblur workflow56. Quality-filtered sequences were clustered into amplicon sequence variants (ASV) and taxonomy was assigned on representative sequences using a Naives Bayes classifier57 trained on the Silva database v13858. The hierarchical structure of the taxonomic classifications from the bacterial communities was visualized with heat trees using the “metacoder” package59 from the R software v3.6.160. Rarefaction curves based on observed ASV and the Good’s coverage index61 were used to evaluate the adequacy of the sampling depth, which was established at 5834 quality-filtered reads per sample. Diversity analyses were performed via the q2-diversity plugin with α-diversity metrics determined using observed ASV, Pielou’s evenness62, and Shannon–Weiner index63 and β-diversity calculated using the weighted UniFrac distances. Visualization of the weighted UniFrac distances was done using the principal coordinate analysis (PCoA) plot.
Statistical analysis
The statistical analyses were conducted using the R software. Prior to analysis, normality was confirmed using the Shapiro–Wilk test (P ≥ 0.13) for dry matter intake (DMI), milk yield, plasma AA, N balance and Neff data. A student t-test was used to compare the aforementioned variables between the low and high Neff groups. Analyses for DMI and milk yield and composition were performed on the collection period data when milk samples were taken (3 days). α-diversity indices were compared using the Kruskal–Wallis test and β-diversity was analyzed with a permutational multivariate ANOVA (PERMANOVA) using the adonis function of the “vegan” package. Differentially abundant ASV within the rumen and fecal bacteriomes between Neff phenotypes were identified using the linear discriminant analysis effect size (LEfSe) implemented in the Galaxy server v1.0 (https://huttenhower.sph.harvard.edu/galaxy/) with default settings. Pearson correlation was used to assess the relationship between individual plasma AA and Neff, while the Spearman’s rank correlation was used to assess the relationship between differentially abundant ASV from the rumen and feces and Neff. The P-values were adjusted for multiple testing using the false discovery rate (FDR) method. Correlation coefficients between 0.51 and 0.70 (− 0.51 and − 0.70), and between 0.71 and 1.00 (− 0.71 and − 1.00) were defined as moderate and strong, respectively64. Statistical significance was declared at P ≤ 0.05.
Ethics approval and consent to participate
All animal procedures were approved by the Mississippi State University Institutional Animal Care and Use Committee (IACUC #19-581).
Data availability
Raw sequences are available at the NCBI Sequence Read Archive (SRA) under the accession no. SRP400507. Detailed information of the complete bioinformatic analysis containing the metadata file and sequential R markdown files with the scripts and commands used in this study can be found at https://github.com/pazlabgit/Neff_holstein_pipeline.
Abbreviations
- AA:
- Amino acids
- ADF:
- Acid detergent fiber
- ASV:
- Amplicon sequence variants
- BW:
- Body weight
- CP:
- Crude protein
- DM:
- Dry matter
- DMI:
- Dry matter intake
- EAA:
- Essential amino acids
- H2SO4 :
- Sulfuric acid
- iADF:
- Indigestible acid detergent fiber
- LEfSe:
- Linear discriminant analysis effect size
- NCBI:
- National center for biotechnology information
- NDF:
- Neutral detergent fiber
- NEAA:
- Non-essential amino acids
- Neff:
- Nitrogen efficiency
- NH3 :
- Ammonia
- PCoA:
- Principal coordinate analysis
- PCR:
- Polymerase chain reaction
- PERMANOVA:
- Permutational multivariate analysis of variance
- QIIME 2:
- Quantitative insights into microbial ecology 2
- SCC:
- Somatic cell count
- SD:
- Standard deviation
- SEM:
- Standard error of the mean
- SRA:
- Sequence read archive
- TEAA:
- Total essential amino acids
- TMR:
- Totally mixed ration
- VFA:
- Volatile fatty acids
References
-
FAOSTAT. Crops and livestock products (2022). https://www.fao.org/faostat/en/#data/QCL. Accessed 1 Oct 2022.
-
USDA-NASS. 2017 Census of Agriculture—United States Data (2019). https://www.nass.usda.gov/Publications/AgCensus/2017/index.php#full_report. Accessed 1 Oct 2022.
-
von Keyserlingk, M. A. G. et al. Invited review: Sustainability of the US dairy industry. J. Dairy Sci. 96, 5405–5425 (2013).
-
Spek, J. W., Dijkstra, J., van Duinkerken, G., Hendriks, W. H. & Bannink, A. Prediction of urinary nitrogen and urinary urea nitrogen excretion by lactating dairy cattle in northwestern Europe and North America: A meta-analysis. J. Dairy Sci. 96, 4310–4322 (2013).
-
Johnson, A. C. B., Reed, K. F. & Kebreab, E. Short communication: Evaluation of nitrogen excretion equations from cattle. J. Dairy Sci. 99, 7669–7678 (2016).
-
USDA-ERS. Milk cost of production estimates (2022). https://www.ers.usda.gov/data-products/milk-cost-of-production-estimates/. Accessed 7 Oct 2022.
-
Powell, J. M., Gourley, C. J. P., Rotz, C. A. & Weaver, D. M. Nitrogen use efficiency: A potential performance indicator and policy tool for dairy farms. Environ. Sci. Policy 13, 217–228 (2010).
-
Fadul-Pacheco, L. et al. Nitrogen efficiency of eastern Canadian dairy herds: Effect on production performance and farm profitability. J. Dairy Sci. 100, 6592–6601 (2017).
-
Gourley, C. J. P., Aarons, S. R. & Powell, J. M. Nitrogen use efficiency and manure management practices in contrasting dairy production systems. Agric. Ecosyst. Environ. 147, 73–81 (2012).
-
Bach, A., Calsamiglia, S. & Stern, M. D. Nitrogen metabolism in the rumen. J. Dairy Sci. 88, E9–E21 (2005).
-
NRC. Nutrient Requirements of Dairy Cattle. 7th rev. edn. (National Academies Press, 2001).
-
Paz, H. A. et al. Rumen bacterial community structure impacts feed efficiency in beef cattle. J. Anim. Sci. 96, 1045–1058 (2018).
-
Wallace, R. J. et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 5, eaav8391 (2019).
-
Ribeiro, G. O. et al. Repeated inoculation of cattle rumen with bison rumen contents alters the rumen microbiome and improves nitrogen digestibility in cattle. Sci. Rep. 7, 1276 (2017).
-
Hoover, W. H. Digestion and absorption in the hindgut of ruminants. J. Anim. Sci. 46, 1789–1799 (1978).
-
Myer, P. R., Wells, J. E., Smith, T. P. L., Kuehn, L. A. & Freetly, H. C. Microbial community profiles of the colon from steers differing in feed efficiency. Springerplus 4, 1–13 (2015).
-
Monteiro, H. F. et al. Rumen and lower gut microbiomes relationship with feed efficiency and production traits throughout the lactation of Holstein dairy cows. Sci. Rep. 12, 1–14 (2022).
-
Castillo, A. R., Kebreab, E., Beever, D. E. & France, J. A review of efficiency of nitrogen utilisation in lactating dairy cows and its relationship with environmental pollution. J. Anim. Feed Sci. 9, 1–32 (2000).
-
Yan, T., Frost, J. P., Agnew, R. E., Binnie, R. C. & Mayne, C. S. Relationships among manure nitrogen output and dietary and animal factors in lactating dairy cows. J. Dairy Sci. 89, 3981–3991 (2006).
-
Colmenero, J. J. O. & Broderick, G. A. Effect of dietary crude protein concentration on milk production and nitrogen utilization in lactating dairy cows. J. Dairy Sci. 89, 1704–1712 (2006).
-
Paz, H. A. & Weiss, W. P. Use of manure concentrations of ash or specific minerals and nitrogen to estimate loss of volatile nitrogen from manure incubated under laboratory conditions. Anim. Feed Sci. Technol. 171, 117–127 (2012).
-
Weiss, W. P. et al. Varying forage type, metabolizable protein concentration, and carbohydrate source affects manure excretion, manure ammonia, and nitrogen metabolism of dairy cows. J. Dairy Sci. 92, 5607–5619 (2009).
-
Bergman, E. N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590 (1990).
-
Seymour, W. M., Campbell, D. R. & Johnson, Z. B. Relationships between rumen volatile fatty acid concentrations and milk production in dairy cows: A literature study. Anim. Feed Sci. Technol. 119, 155–169 (2005).
-
Paz, H. A. et al. Ruminal degradation and intestinal digestibility of protein and amino acids in high-protein feedstuffs commonly used in dairy diets. J. Dairy Sci. 97, 6485–6498 (2014).
-
Wang, L. et al. Bacterial community diversity associated with different utilization efficiencies of nitrogen in the gastrointestinal tract of goats. Front. Microbiol. 10, 239 (2019).
-
Toral, P. G. et al. Comparison of ruminal lipid metabolism in dairy cows and goats fed diets supplemented with starch, plant oil, or fish oil. J. Dairy Sci. 99, 301–316 (2016).
-
Zhang, J. et al. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front. Microbiol. 8, 2206 (2017).
-
Patton, R. A., Hristov, A. N., Parys, C. & Lapierre, H. Relationships between circulating plasma concentrations and duodenal flows of essential amino acids in lactating dairy cows. J. Dairy Sci. 98, 4707–4734 (2015).
-
Chaudhry, F. A., Reimer, R. J. & Edwards, R. H. The glutamine commute: Take the N line and transfer to the A. J. Cell Biol. 157, 349 (2002).
-
Myer, P. R., Smith, T. P. L., Wells, J. E., Kuehn, L. A. & Freetly, H. C. Rumen microbiome from steers differing in feed efficiency. PLoS One 10, e0129174 (2015).
-
Xue, M. Y., Sun, H. Z., Wu, X. H., Guan, L. L. & Liu, J. X. Assessment of rumen bacteria in dairy cows with varied milk protein yield. J. Dairy Sci. 102, 5031–5041 (2019).
-
Stevenson, D. M. & Weimer, P. J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75, 165–174 (2007).
-
Henderson, G. et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5, 14567 (2015).
-
Xue, M. Y., Sun, H. Z., Wu, X. H., Liu, J. X. & Guan, L. L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 8, 1–19 (2020).
-
Mizrahi, I., Wallace, R. J. & Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566 (2021).
-
Stanton, T. B. & Canale-Parola, E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch. Microbiol. 127, 145–156 (1980).
-
Mao, S., Zhang, M., Liu, J. & Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 5, 16116 (2015).
-
Lourenco, J. M. et al. Fecal microbiome differences in Angus steers with differing feed efficiencies during the feedlot-finishing phase. Microorganisms 10, 1128 (2022).
-
Boutard, M. et al. Functional diversity of carbohydrate-active enzymes enabling a bacterium to ferment plant biomass. PLoS Genet. 10, 1004773 (2014).
-
Otter, A. & Uzal, F. A. Clostridial diseases in farm animals: 1. Enterotoxaemias and other alimentary tract infections. In Pract. 42, 219–232 (2020).
-
AOAC International. Official Methods of Analysis 17th edn. (AOAC International, 2000).
-
Van Soest, P. J., Robertson, J. B. & Lewis, B. A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597 (1991).
-
Tebbe, A. W. & Weiss, W. P. Evaluation of creatinine as a urine marker and factors affecting urinary excretion of magnesium by dairy cows. J. Dairy Sci. 101, 5020–5032 (2018).
-
Huhtanen, P., Kaustell, K. & Jaakkola, S. The use of internal markers to predict total digestibility and duodenal flow of nutrients in cattle given six different diets. Anim. Feed Sci. Technol. 48, 211–227 (1994).
-
Nadeau, E., Englund, J.-E. & Gustafsson, A. H. Nitrogen efficiency of dairy cows as affected by diet and milk yield. Livest. Sci. 111, 45–56 (2007).
-
Nkrumah, J. D. et al. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 84, 145–153 (2006).
-
Paz, H. A., Anderson, C. L., Muller, M. J., Kononoff, P. J. & Fernando, S. C. Rumen bacterial community composition in Holstein and Jersey cows is different under same dietary condition and is not affected by sampling method. Front. Microbiol. 07, 1206 (2016).
-
de La Guardia-Hidrogo, V. M. & Paz, H. A. Influence of industry standard feeding frequencies on behavioral patterns and rumen and fecal bacterial communities in Holstein and Jersey cows. PLoS One 16, e0248147 (2021).
-
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
-
Paz, H. A. & Kononoff, P. J. Lactation responses and amino acid utilization of dairy cows fed low-fat distillers dried grains with solubles with or without rumen-protected lysine supplementation. J. Dairy Sci. 97, 6519–6530 (2014).
-
Le Boucher, J., Charret, C., Coudray-Lucas, C., Giboudeau, J. & Cynober, L. Amino acid determination in biological fluids by automated ion-exchange chromatography: Performance of Hitachi L-8500A. Clin. Chem. 43, 1421–1428 (1997).
-
Fekkes, D. State-of-the-art of high-performance liquid chromatographic analysis of amino acids in physiological samples. J. Chromatogr. B Biomed. Appl. 682, 3–22 (1996).
-
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
-
Bokulich, N. A. et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59 (2013).
-
Amir, A. et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2, e00191-16 (2017).
-
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018).
-
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
-
Foster, Z. S. L., Sharpton, T. J. & Grünwald, N. J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 13, e1005404 (2017).
-
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2022).
-
Good, I. J. The population frequencies of species and the estimation of population parameters. Biometrika 40, 237–264 (1953).
-
Pielou, E. C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 13C, 131–144 (1966).
-
Shannon, C. E. A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423 (1948).
-
Mukaka, M. A guide to appropriate use of Correlation coefficient in medical research. Malawi Med. J. 24, 69 (2012).
Acknowledgements
The authors thank Kenneth Graves and Colby Hardin from the Bearden Dairy Research Center for their support with daily cow management and sample collection.
Ethics declarations
Competing interests
The authors declare no competing interests
https://www.nature.com/articles/s41598-023-37891-7